The starting point for the extra credit assignment are the results from your BLASTP searches with the predicted proteins of your ab initio analysis (exercise 7B). (If you got 'no significant matches' for a given predicted protein, there is no need to go further with that predicted protein, since it likely is not real. Include this information, however, in the table as shown in the 'products' section below.)
Search among your matches for a C. elegans (not just any Caenorhabditis) gene, then put this gene into a search at WormbaseYou might also find this site useful to get information about protein functions: KEGG: Kyoto Encyclopedia of Genes and Genomes
As a 'genome annotator,' one of your tasks is to make an educated guess about the function of genes you have uncovered via your ab initio and homology (BLAST) analyses. The simplest/ easiest way is if you can identify a well-studied ortholog in another organism. An ortholog is essentially the 'same gene' in another organism, and will typically yield a high blast score (and low E-value) in comparisons.
Since the genomic sequence analyzed is from Oscheius myriophila, a nematode relative of the very well-studied nematode model organism C. elegans, if you can identify a C. elegans ortholog, you have a reasonable chance of identifying a likely function for that predicted gene - if this is one of the many genes already characterized in C. elegans. Look through your BLASTP blast hits to find the highest scoring C. elegans gene. [One way to speed up this process is to re-do the BLASTP search and restrict the organism to C. elegans. But also pay attention to your original 'nr' database search, as should be clear from Example 2 below.]
C. elegans gene names that you can search in Wormbase (top level, default search 'for a gene') will have either this form: cat-1 (3 or 4 letters, dash, number) and/or W01C8.6 (e.g., capital letter(s), numbers, letter, number, period, number).
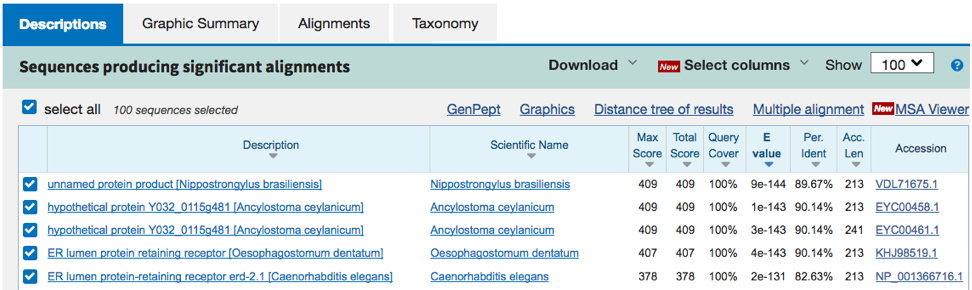
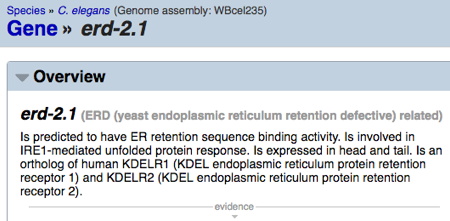
Example 1. From this BLASTP search (results below), there is a clear C. elegans ortholog (bottom line). The entire query sequence is aligned (Query Cover = 100%), and E-value very low (e-131). The C. elegans gene name is erd-2.1; when searched in Wormbase, this yields information (below) that the sequence is orthologous to the human KDELR1 - an 'ER protein retention receptor' (a reasonable descriptor for the table entry 'Type of protein encoded').

Wormbase search for erd-2.1 result (below):
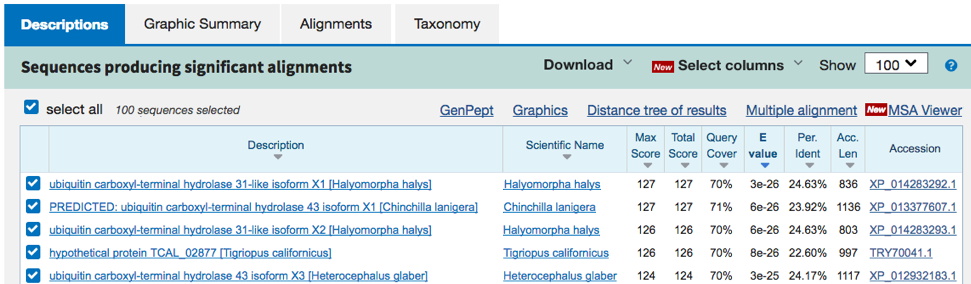
Example 2. Interestingly, in this search (results below), the best matches (E values ~ e-26) are to proteins in insects and mammals. The best C. elegans matches have much higher E-values (e-10). This means the C. elegans homolog is not that similar, and definitely not an ortholog. But examination of the gene names / descriptions in both cases make it clear our Omy predicted protein is somewhat similar to 'ubiquitin carboxyl-terminal hydrolase-like' proteins (but only ~24% identical over a portion of the query sequence). If you search the named gene 'usp-48' in Wormbase, the description is of a 'ubiquitinyl hydrolase' or a 'ubiquitin-specific protease.' Any of those three descriptions would be acceptable for the table entry for 'Type of protein encoded.'
Search with 'nr' database
Search restricted to C. elegans
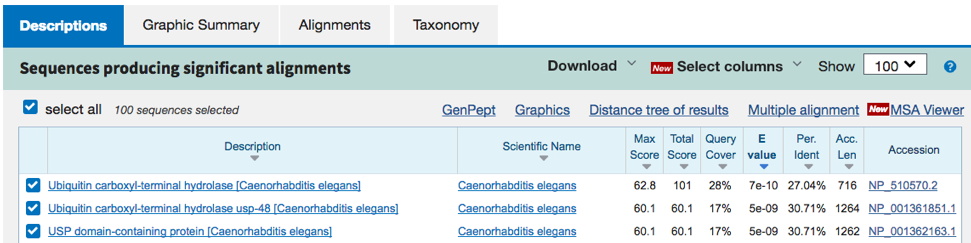
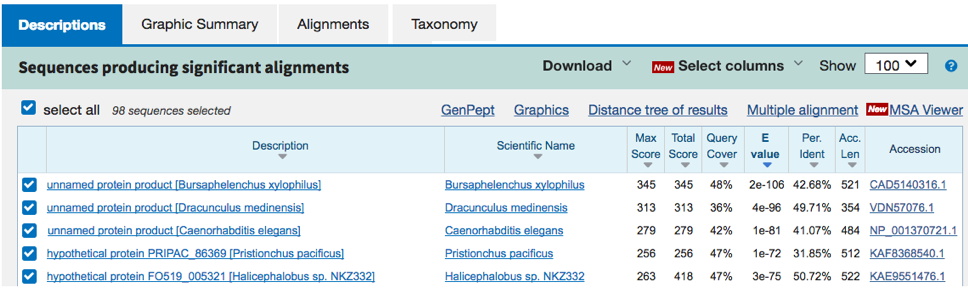
Example 3. Sometimes you get a result with no information about the identity of the predicted protein (see below - all 'unamed protein product' or 'hypothetical protein' matches). The predicted protein is likely real (actually exists) since there are strong matches in many other species (even orthologs), but no one has studied the predicted protein yet, and the AA sequence is novel enough that there are no clues yet to its function. In this case, simply enter something like 'unnamed / hypothetical protein' for the type of protein encoded. This type of result is becoming rare, as there may be at least some prediction of specific protein domains within a given sequence, providing a minimal clue to function.
Information about possible gene function - distill your analysis of BLASTP results to this simple additional table (example below).
- only for genes with BLASTP and/or BLASTX matches, but include all FGENESH predicted genes in the table - e.g. see genes 3, 5. The gene numbering matches up with the main analysis table.
| Gene | C. elegans homolog | Type of protein encoded |
| 1 | cat-1 gene | Vesicular monoamine transporter |
| 2 | msh-1 / H26D21.2 | Mismatch repair protein MutS family |
| 3 | none / NA | no significant matches |
| 4 | ZK550.3 | Oligopeptidase |
| 5 | none / NA | no significant matches |
| 6 | etc. | etc. |
| * | none | WD40 repeat protein |