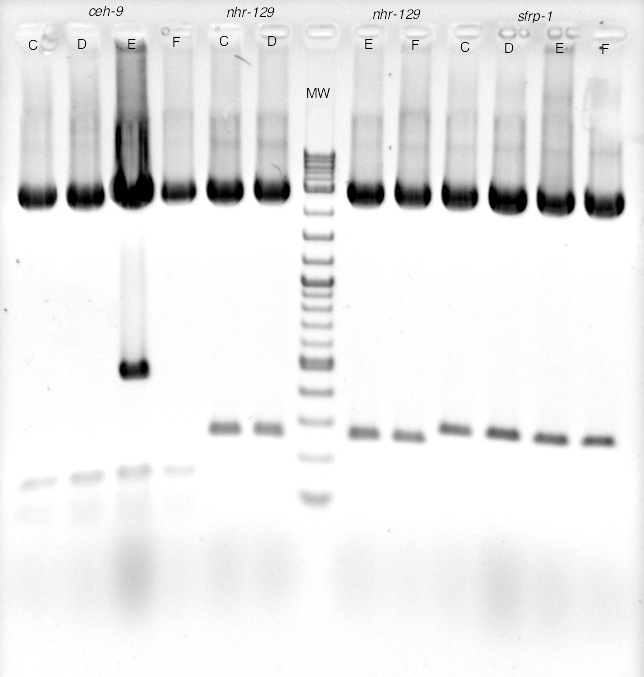
Some transformations yielded colonies that when respread, cultured, and plasmid DNA purified gave the expected result - plenty of plasmid DNA. Others did not. To find out what happened, we investigated further.
Nine different clones from transformation respreads by various students were grown as overnight cultures, and DNA purified by a plasmid miniprep. The results from these fell into two distinct classes (designated category A and B). DNA concentrations for the two categories are shown at right.