Get your protein sequences in FASTA format.
Be sure to give each protein a short, useful name in the first line of the FASTA format sequence. Start with a three letter abbreviation for the species binomen (One capital letter for the genus, followed by two species name letters. E.g., 'Cel' for C. elegans; 'Ppa' for P. pacificus.) Keep track of the full species name in a separate file ((Example species + protein list)).
For example, perhaps you have a file titled 'ppa_stranded_DN26056_c0_g2_i4' that is a likely ortholog of SFRP-1 from P. pacificus - call it '>Ppa_SFRP-1'
As noted in the week 2 instructions, put your protein sequences in the following order:
Cel
Ppa
Pgi
Mja
Asu (likely 2 proteins)
Dme
Hsa
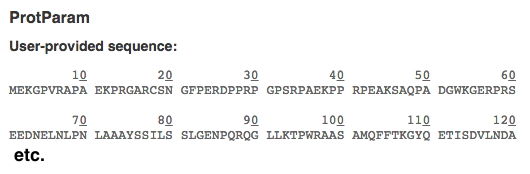
[If you need a numbered version of your key POI, use the ExPASy protein parameters tool to get a nicely numbered version that will make it easier to find specific AAs. It will appear at the top of the ProtParam result. Example of a numbered sequence shown below:

1B. Paste POI Sequences into a single FASTA format file
(Example FASTA file + species list)
Making alignments of related protein sequences is an important bioinformatic technique for a variety of purposes. Alignments show what portions of a molecule are shared between two or more molecules.
Parts of a given protein that are highly conserved (that is, unchanged over evolutionary time) in many distantly-related organisms are likely to be important functional regions. In other words, that portion of the protein cannot be altered by mutations without destroying or reducing the protein's function. Organisms with such mutations are selected against - out-competed for reproductive success - so the changes are lost. On the other hand, regions of the protein (or specific amino acids) that can change without disrupting protein function will evolve - randomly change over time - and be different in distantly-related organisms.
Run the alignment with Clustal Omega.
The program we will use for alignment is Clustal Omega and is found at the European Bioinformatics Institute (EBI) website. There are other multiple sequence aligmnent programs available, including on the NCBI website.
Under 'STEP 1,' paste the collected sequences into the large window.
Under 'STEP 2,' click on 'More options...' to change this parameter:
ORDER (last among all the options): change default 'aligned' to 'input' - this keep your key POI sequence (e.g., the Ppa sequence) at the top of the MSA.
Part of an example alignment with this format (using the GCH1 proteins file) is shown below:

You might like seeing and saving a colorful version - click 'Show Colors'. It does provide some additional information.
Click 'Phylogenetic Tree' to see a tree generated using this alignment.
If you get a poor alignment, consider removing a protein sequence that may be disrupting the alignment. Sometimes a protein you've chosen is quite different from all the others.
Files you can use for demonstration purposes: Example FASTA files