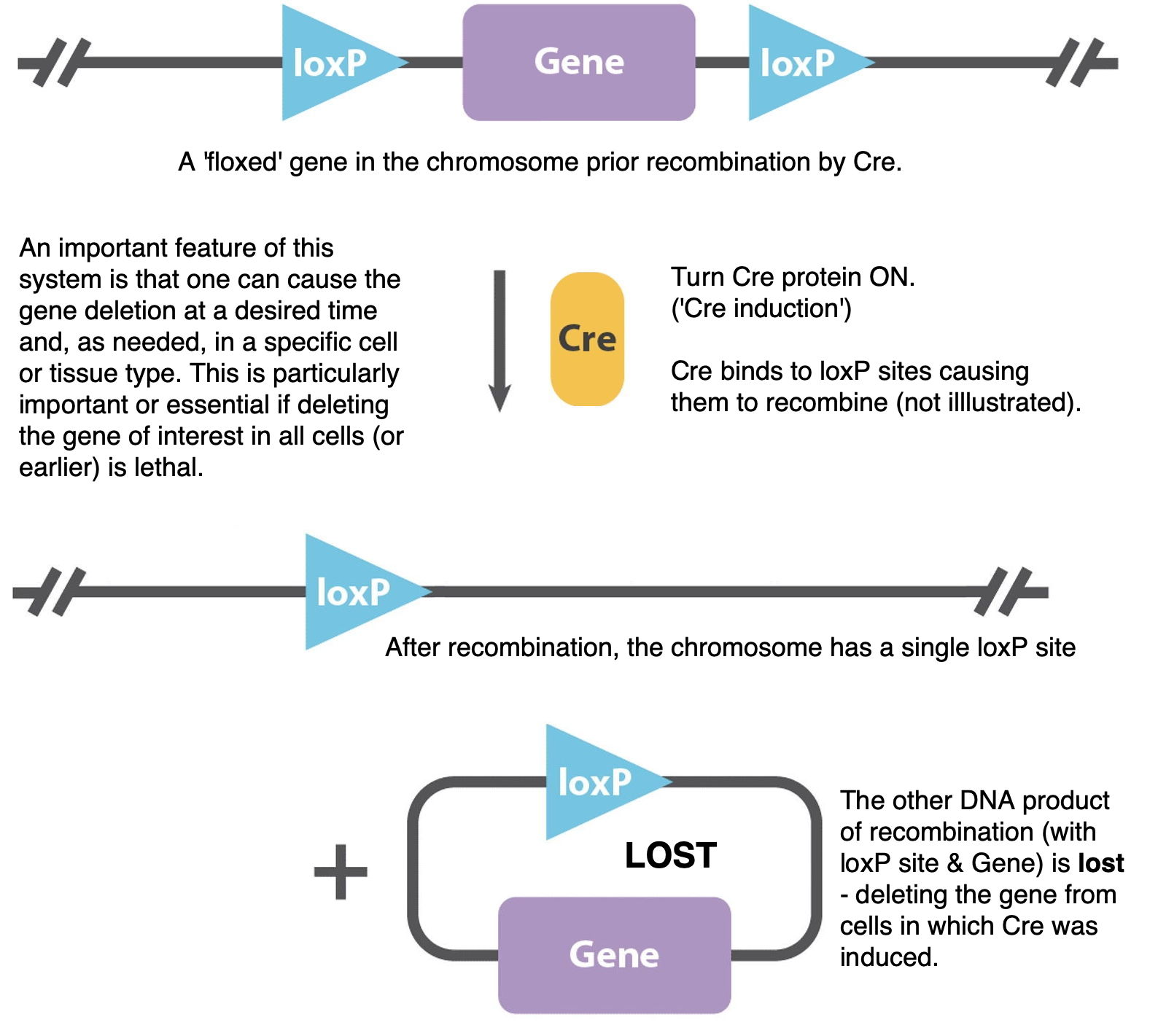
For example, what is important about using the Cre-Lox system is that it allows one to delete a gene at a desired time, and if set up correctly, only in a desired tissue or cell type. [Note that Cre-Lox can be used to manipulate the genome in other ways as well.]
In the Reed paper, this allows the scientists to delete the APC gene in adult mice specfically in liver cells or in intestinal cells. They can also delete the beta-catenin gene or the c-Myc gene. They can delete more than one gene to create double mutants. That's why they are using the method.
Near the bottom of the page is a more detailed explanation of how the Cre-Lox system is used to do this.
Ki67 - a Ki67 antibody marks a nuclear protein found only in proliferating cells and not in quiescent cells. So, it provides similar information to BrdU.
IHC - immunohistochemistry - here, using an antibody to a given protein, then an enzyme-linked secondary antibody. The enzyme substrate DAB makes a dark brown precipitate indicating presence of the protein of interest (i.e., b-catenin, c-Myc, Ki67)
Microarray analysis here involves comparing RNA expression levels from many genes in different genotypes. [Microarray analysis has largely been superceded by RNA-Seq - high-throughput sequencing of millions of RNA molecules to analyze expression of all genes.]
SAM (Significance Analysis of Microarrays) is a standard statistical method for determining the statistical significance of differences in gene expression in microarray expression data - always comparing two different conditions (such as the expression of a specific gene in two different genotypes).
Quantitative PCR - (in this context for the Reed paper, qRT-PCR) - method for examining gene expression of one gene or a few genes at a time (in contrast to microarray analysis, which simultaneously analyzes the expression of hundreds to thousands of genes). Given the scale and potential 'noisiness' of microarray analysis, qRT-PCR is frequently used to confirm results of the microarray analysis for a few specific genes.
Cre-Lox system terminology & explanations - how the authors deleted genes
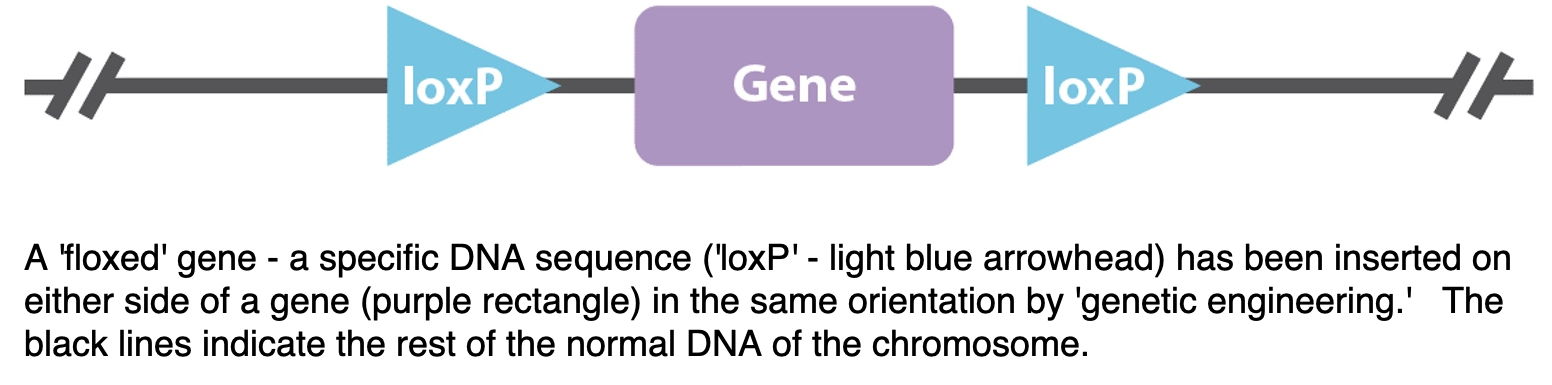
floxed - 'flanked by loxP sites'

A 'floxed' gene is one that has engineered 'loxP' recombination sites on the left and right ('flanking'). [Illustrated above; the entire gene deletion process is illustrated below.] When the Cre recombinase protein is expressed, the Lox sites recombine to either remove the DNA between them (lox sites in same orientation) or invert the segment of DNA (lox sites in opposite orientations). In the first case, this makes a deletion allele of the gene in question - but only in the cells expressing the Cre protein. (For example, this can be used to avoid a lethal phenotype in a regular homozygous deletion mutant. Or, to study the mutant phenotype only in a specific tissue.) The Cre recombinase protein can be turned on in specific cell or tissue types. In the Reed et al., 2008 paper, Cre recombinase protein is induced (expression turned on) by beta-naphthoflavone injection.
For further information:
A site that explains a Cre-Lox transgenics system: The Cre/loxP recombination system in transgenic mice
The Wikipedia entry is also decent: floxed
See 'Conditional targeting' on this Mouse transgenics site (you may need to read more of the site to understand this section):