 We have found recently that the original allele of bas-1, ad446, is a ~4300 bp deletion that removes almost the entire coding regions of both C05D2.4 and C05D2.3 predicted genes.
We have found recently that the original allele of bas-1, ad446, is a ~4300 bp deletion that removes almost the entire coding regions of both C05D2.4 and C05D2.3 predicted genes.Wormbase gene report for bas-1
bas-1 alleles: ad446, n2948, n3008, pa4, tm351(deletion mutant, not fully characterized yet)
The gene name bas-1 stands for biogenic amine synthesis abnormal/related. The original allele (ad446) was isolated by Gian Garriga in an Egl mutant screen. Ironically, however, when the egl mutant was recovered in an out-cross, the serotonin-deficiency phenotype had disappeared. ad446 was apparently a second mutation in the background of the original strain. The mutation does not cause an obvious Egl phenotype (although it does have subtle effects on egg laying behavior (see below).
bas-1 mutants are serotonin- and dopamine-deficient (Loer & Kenyon, 1993; Sawin & Horvitz, unpublished - cited in Sawin et al., 2000) by serotonin-immunoreactivity (serotonin-IR) and formaldehyde induced flourescence (FIF, for dopamine). A low level of serotonin-IR has been reported in bas-1(ad446) (Weinshenker et al., 1995); we have subsequently found that the bas-1 mutant alleles ad446, n2948, n3008, and pa4 have similar low levels of serotonin-IR (analysis pending on a new deletion allele, tm351). We are uncertain how this correlates with actual serotonin levels in the organism.
Behaviorally, bas-1 mutants are defective in male turning behavior (Loer & Kenyon, 1993), starvation-induced enhanced slowing (Sawin et al., 2000), and induction of active-state egg-laying (Waggoner et al., 1998). Other reported behavioral abnormalities of bas-1 mutants include "area-restricted search" deficiency (Hill et al., 2000).
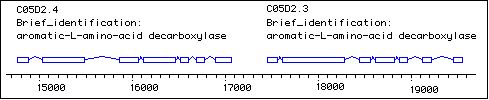
bas-1 mutants can be restored to wild type serotonin-IR by treatment with 5HT (serotonin), but not with its immediate precursor, 5HTP (Loer & Kenyon, 1993). This is consistent with a loss of aromatic amino acid decarboxylase activity. bas-1 maps genetically between dpy-17 and unc-32 on chromosome III. Genome sequence revealed the presence of two adjacent aromatic amino acid decarboxylases (C05D2.4, C05D2.3) in this interval. The cosmid C05D2 and 15.5 kb subclone C05D2XN (courtesy Fred Wolf & Gian Garriga) contains these AAADC genes and rescues the serotonin deficiency of bas-1 mutants (ref a, ref b). We have found that just one these sequences (C05D2.4) is necessary to rescue bas-1 mutants; furthermore alleles n2948, n3008 and pa4 all contain point mutations in the C05D2.4 coding sequence (Hare & Loer, 2000).  We have found recently that the original allele of bas-1, ad446, is a ~4300 bp deletion that removes almost the entire coding regions of both C05D2.4 and C05D2.3 predicted genes.
We have found recently that the original allele of bas-1, ad446, is a ~4300 bp deletion that removes almost the entire coding regions of both C05D2.4 and C05D2.3 predicted genes.
We have yet to determine the function of C05D2.3, which is located immediately downstream of C05D2.4. The close spacing of the genes suggests they may be expressed as an operon, however, there is no evidence for this to date. Furthermore, expression profiles analyzed from microarray hybridization experiments do not support the idea that these genes constitute an operon (Tom Blumenthal, personal communication). The presence of cDNAs from both sequences in the EST database and our own RT-PCR experiments show that both genes are expressed. The existing C05D2.3 cDNA is partial; it has not yet been shown to be spliced to either SL1 or SL2. C05D2.4 is spliced to SL1;  we have also found three different alternatively spliced forms of C05D2.4 by RT-PCR (E. Hare & Loer, unpublished).
we have also found three different alternatively spliced forms of C05D2.4 by RT-PCR (E. Hare & Loer, unpublished).
 |
Published References (unpublished references above are linked to the abstract)
Loer C, Kenyon C (1993) Serotonin-deficient mutants and male mating behavior in the nematode Caenorhabditis elegans. J. Neurosci. 13:5407-5417.
Sawin ER, Ranganathan R, Horvitz HR (2000) C. elegans locomotory rate is modulated by the environment through a dopaminergic pathway and by experience through a serotonergic pathway. Neuron 26:619-631.
Waggoner LE, Zhou GT, Schafer RW, Schafer WR (1998) Control of alternative behavioral states by serotonin in Caenorhabditis elegans. Neuron 21: 203-214.
Weinshenker D, Garriga G, Thomas JH (1995) Genetic and pharmacological analysis of neurotransmitters controlling egg laying in C. elegans. J. Neurosci. 15: 6975-6985.