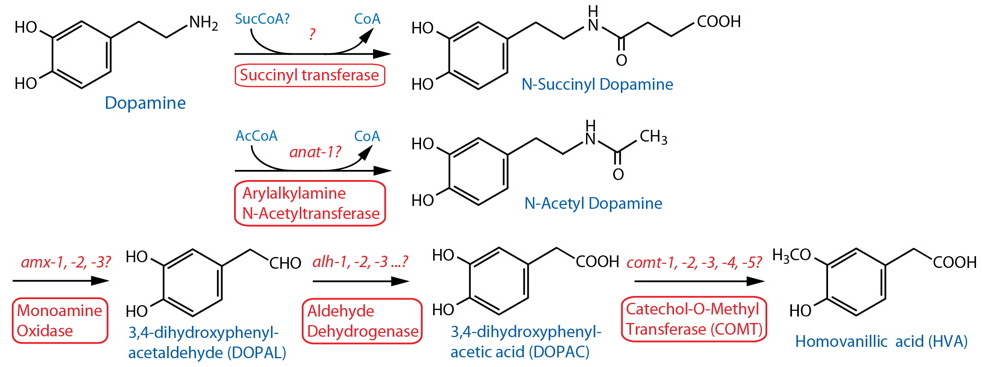
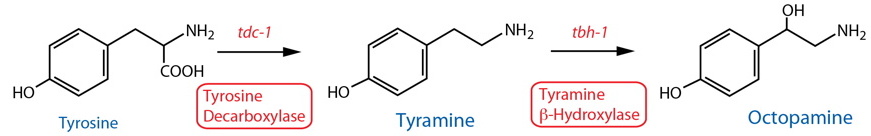
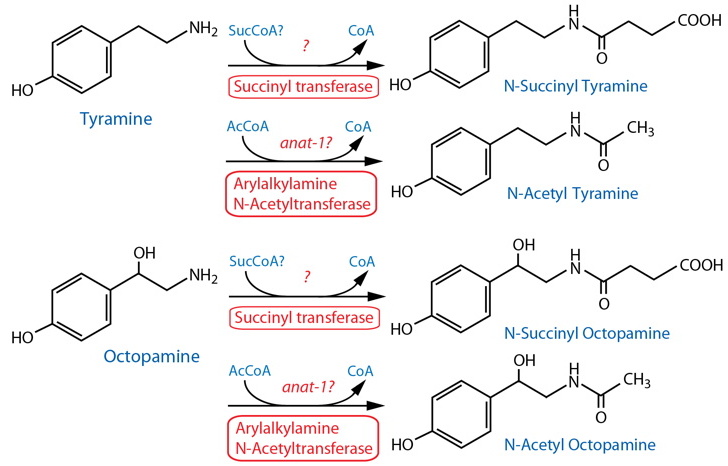
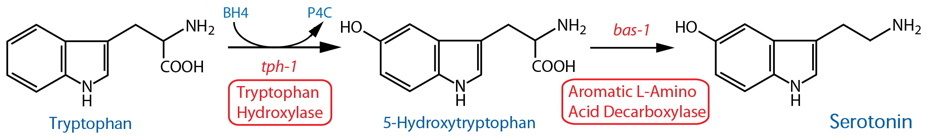
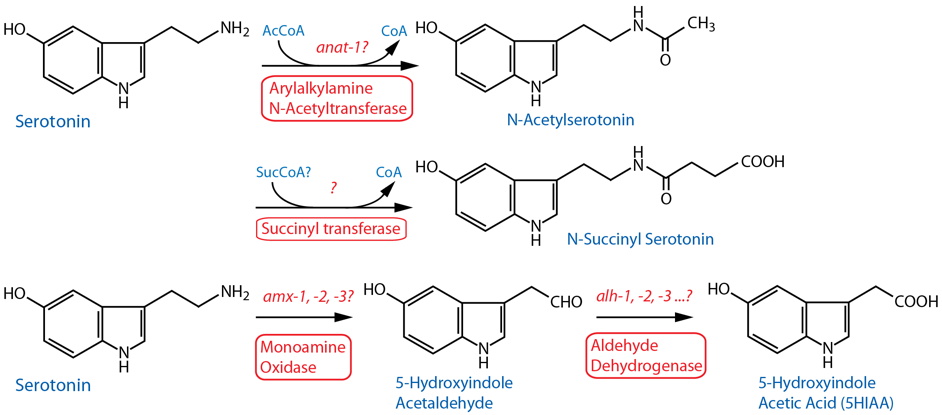
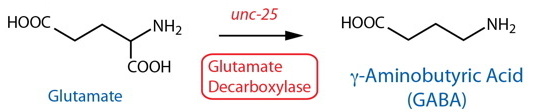
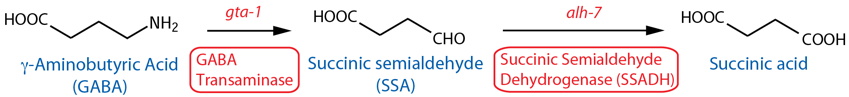
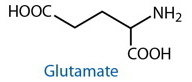
DESCRIPTION
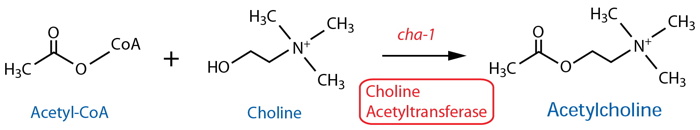
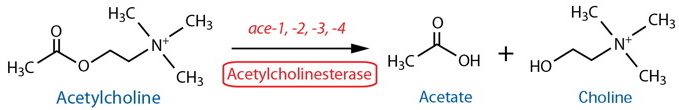
By Class:
ADF, AIA, AIM, AIN, AIY, ALN, AS, ASJ, AVA, AVB, AVD, AVE, AVG, AWB, CA(m), CEM(m), DA, DB, DVA, DVE(m), DX1(m), DX2(m), HOB(m), HSN(h), I1, I3, IL2, M1, M2, M4, M5, MC, Â PCB(m), PCC(m), PDA, PDB, PDC(m), PGA(m), PLN, PVC, PVN, PVP, PVV(m), PVX(m), PVY(m), PVZ(m), R1A(m), R2A(m), R3A(m), R4A(m), R6A(m), RIB, RIF, RIH, RIR, RIV, RMD, RMF, RMH, SAA, SAB, SDQ, SIA, SIB, SMB, SMD, SPC(m), SPV(m), URA, URB, URX, VA, VB, VC(h)
By Class (with numbers and notes):
ADF(2), AIA(2), AIM(2)4, AIN(2), AIY(2), ALN(2), AS1-11, ASJ(2), AVA(2), AVB(2), AVD(2), AVE(2), AVG5, AWB(2), CA1-9(m)4, CEM(4, m), DA1-9, DB1-7, DVA6, DVE(m), DX1(m)7, DX2(m)7, HOB(m), HSN(2, h), I1(2)5, I35, IL2(6), M15, M2(2)5, M4, M5, MC(2)5, PCB(2, m), PCC(2, m), PDA, PDB, PDC(m)8, PGA(m)8, PLN(2), PVC(2), PVN(2)5, PVP(2), PVV(m), PVX(m), PVY(m), PVZ(m), R1A(2, m), R2A(2, m), R3A(2, m), R4A(2, m), R6A(2, m), RIB(2)8, RIF(2), RIH, RIR, RIV(2), RMD(6), RMF(2), RMH(2), SAA(4)5, SAB(3), SDQ(2), SIA(4), SIB(4), SMB(4), SMD(4), SPC(2, m), SPV(2, m), URA(4), URB(2), URX(2), VA1-12, VB1-11, VC1-6(h)
Head: ADF(2), AIA(2), AIM(2)4, AIN(2), AIY(2), AS1, ASJ(2), AVA(2), AVB(2), AVD(2), AVE(2), AVG5, AWB(2), CEM(4, m), DA1, DB1-2, IL2(6), RIB(2)8, RIF(2), RIH, RIR, RIV(2), RMD(6), RMF(2), RMH(2), SAA(4)5, SAB(3), SIA(4), SIB(4), SMB(4), SMD(4), URA(4), URB(2), URX(2), VA1, VB1-2
Pharynx: I1(2)5, I35, M15, M2(2)5, M4, M5, MC(2)5
Ventral nerve cord & body: AS2-10, CAn(1-9)(m)4, DA2-7, DB3-7, HSN(2, h), SDQ(2), VA2-11, VB3-11, VC1-6(h)
Tail: ALN(2), AS11, DA8-9, DVA6, DX1(m)7, DX2(m)7, HOB(m), PCB(2, m), PCC(2, m), PDA, PDB, PDC(m)8, PGA(m)8, PLN(2), PVC(2), PVN(2)5, PVP(2), PVV(m), PVX(m), PVY(m), PVZ(m), R1A(2, m), R2A(2, m), R3A(2, m), R4A(2, m), R6A(2, m), SPC(2, m), SPV(2, m), VA12
Note: Somas found in the retrovesicular ganglion (RVG) are listed in the head; those in the preanal ganglion (PAG) are listed in the tail.