





 |
 |
 |
 |
 |
 |
 |
 |


The evidence for classical neurotransmitters in C. elegans neurons - Local version (2025) / WormAtlas published version (2022 major update)
Criteria for assigning a neurotransmitter function in C. elegans - Local version (2022) / WormAtlas published version (2022)
We have recently begun working in earnest on the nervous system of the nematode 'satellite model organism' Pristionchus pacificus, comparing it with that of C. elegans, for which we have deep knowledge about how nerve cells choose precisely what kind of nerve cell to become - the question of neuronal specification. As a important resource for such work, we are working to characterize the Pristionchus nervous system fully - identifying every single neuron, to match the full description of the C. elegans nervous system.
As a first step in comparing neuronal specification in the two species, we are characterizing the neuronal expression of two transcription factors (TFs) known to control neuronal fate, especially neurotransmitter type, in C. elegans: unc-3 and unc-86 -- the COE-type TF unc-3 specifies many neurons to use acetylcholine (ACh). The POU homeodomain TF unc-86 helps specify some C. elegans serotonergic neurons such as the NSMs found in the pharynx.
[Homologs of these TFs play important roles in the vertebrate nervous system as well.]
This work is in collaboration with the laboratories of Oliver Hobert (Columbia University, NYC) and Ralf Sommer (Max Planck Institute for Developmental Biology, Tübingen, Germany). Work on other genes of interest including the neurotransmitter transporters dat-1 and cat-1 involves collaboration with Ray Hong (CSU, Northridge).
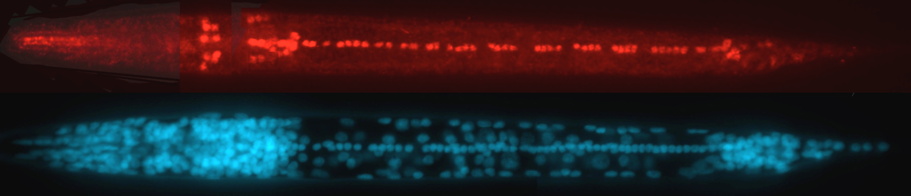
Image below: Ventral view of Pristionchus larva with protein marker-tagged Ppa-unc-3 gene (via CRISPR/Cas9, strain by H. Witte of Sommer lab). Top - neuronal nuclei (red) expressing Ppa-UNC-3 protein. Bottom - same worm showing all nuclei, stained with DNA stain DAPI. The head of the worm (left) contains the 'brain' with >100 neurons, including many expressing unc-3. The ventral nerve cord, also with many neurons expressing unc-3, is apparent on the ventral midline of the body running from the head to the tail (right) which has another cluster of many neurons.
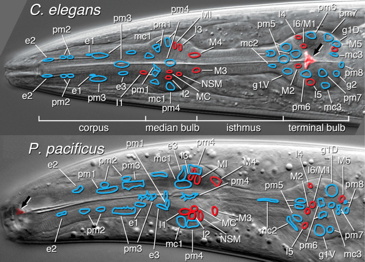
Cells of the pharynx in C. elegans and P. pacificus; neurons indicated in red (Fig. 1 C, D from Bumbarger et al., 2013).
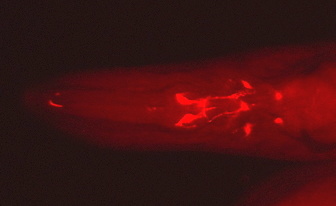
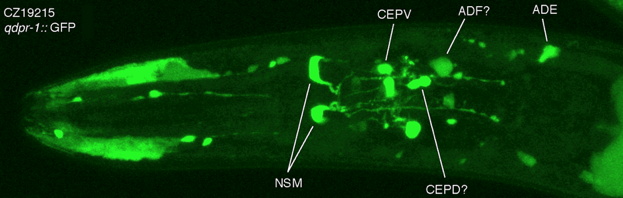
We study the specification, function and patterning of serotonin-using ('serotonergic') neurons in C. elegans. What genes are required to make serotonin and how are they regulated? How are those genes turned on in specific neurons and not in others? How are serotonergic neurons positioned at specific locations in
the animal? We have collaborated with the Wolff lab (Carleton College) to study how sex-specific serotonergic neurons are patterned in the worm.
We also have
ongoing research projects on this topic with students in classes, especially Biology 376 - Animal Development.
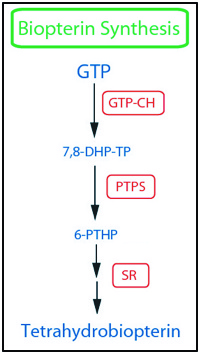
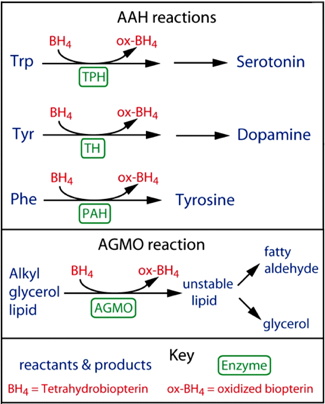
In order to synthesize both serotonin and dopamine, animals require a chemical called tetrahydrobiopterin (BH4) or biopterin. This enzymatic cofactor is usually synthesized by
the cells that need it; therefore, serotonergic and dopaminergic neurons turn on genes encoding biopterin synthetic enzymes. Additional genes required for biopterin use
include those encoding enzymes for biopterin regeneration and a protein that helps regulate biopterin synthesis. We have studied these genes and the proteins they encode in
collaboration with the Chisholm (UCSD) and Werner (Univ. of Innsbruck, Austria) labs.
Like all animals, C. elegans has several enzymes that require biopterin to function. These include three related enzymes, the aromatic amino acid
hydroxylases (AAAHs), and a lipid metabolic enzyme alkyl glycerol monooxygenase (AGMO). This enzyme was characterized in humans relatively recently (2010); we have previously studied
its function in C. elegans in collaboration with the Chisholm, Werner, Martinez (Univ of Bergen, Norway), and Hodgkin (Univ. of Oxford, UK) labs.
We sometimes collect nematodes in the wild (or are given them) and find interesting worms. For example, USD Microbiology students found some worms in the top layer of a Winogradsky column made using San Diego River mud and gave them to us. This fascinating nematode (below) is a Propanagrolaimus sp. that subsequently appeared in a publication with the Schierenberg lab, and soon will have its complete genome sequenced by Philipp Schiffer's lab (Univ of Cologne, Germany). Amazingly, its closest known relative is found in an underground cave river in Romania...
Evolution of Neuronal Specification - "Evo-Devo-Neuro"
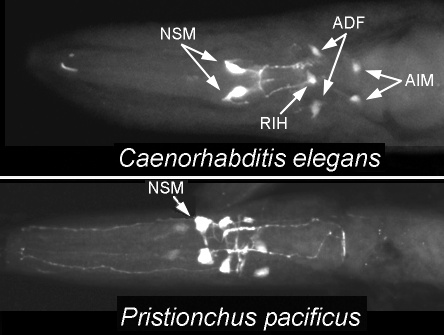
Serotonergic neurons in the heads of C. elegans and P. pacificus (data from Rivard et al., 2010).
Serotonergic Neurons in C. elegans
Biopterin Synthesis Genes
Biopterin-dependent Enzymes
Nematodes from the wild
