1. Find a crystal structure for your protein of interest. Go to NCBI - Structure. (You may also find a direct link to a protein's crystal structure - or multiple structures - from its main 'gene page.')
2. Search for a structure for your protein of interest in the NCBI Structure database with the name of your protein. There may be several different version of structures. Look for an indication of a larger, more complete sequence. Get the PDB ID for the structure. That's what you will use to access the structure below. These identifiers are made of 4 capital letters and numbers. Some examples: 2GK1, 1HD.
[Note: some of these proteins may not have crystal structures for the human protein - there may be a closely related protein (e.g., another mammalian protein like rat instead of human) that you can use for this purpose. Check with the instructor if you need help.]
For transcription factors, try to find a version of the protein bound to DNA. Depending on the TF, this may be as a monomer, a dimer (or other multimeric version) or with a different TF partner.
3. Click on the entry, which will take you to a MMDB Structure Summary
4. View structures using iCn3D - "I see in 3-D" -NCBI WebGL-based Structure Viewer page
When you're ready to use it, click on "OPEN iCn3D" (see image below).

Then enter a PDB file number by clicking FILE, and 'Retrieve by ID' - PBD ID, and wait for the image to load. The first time the program loads, this may take a while. Then you can view it in a variety of ways.
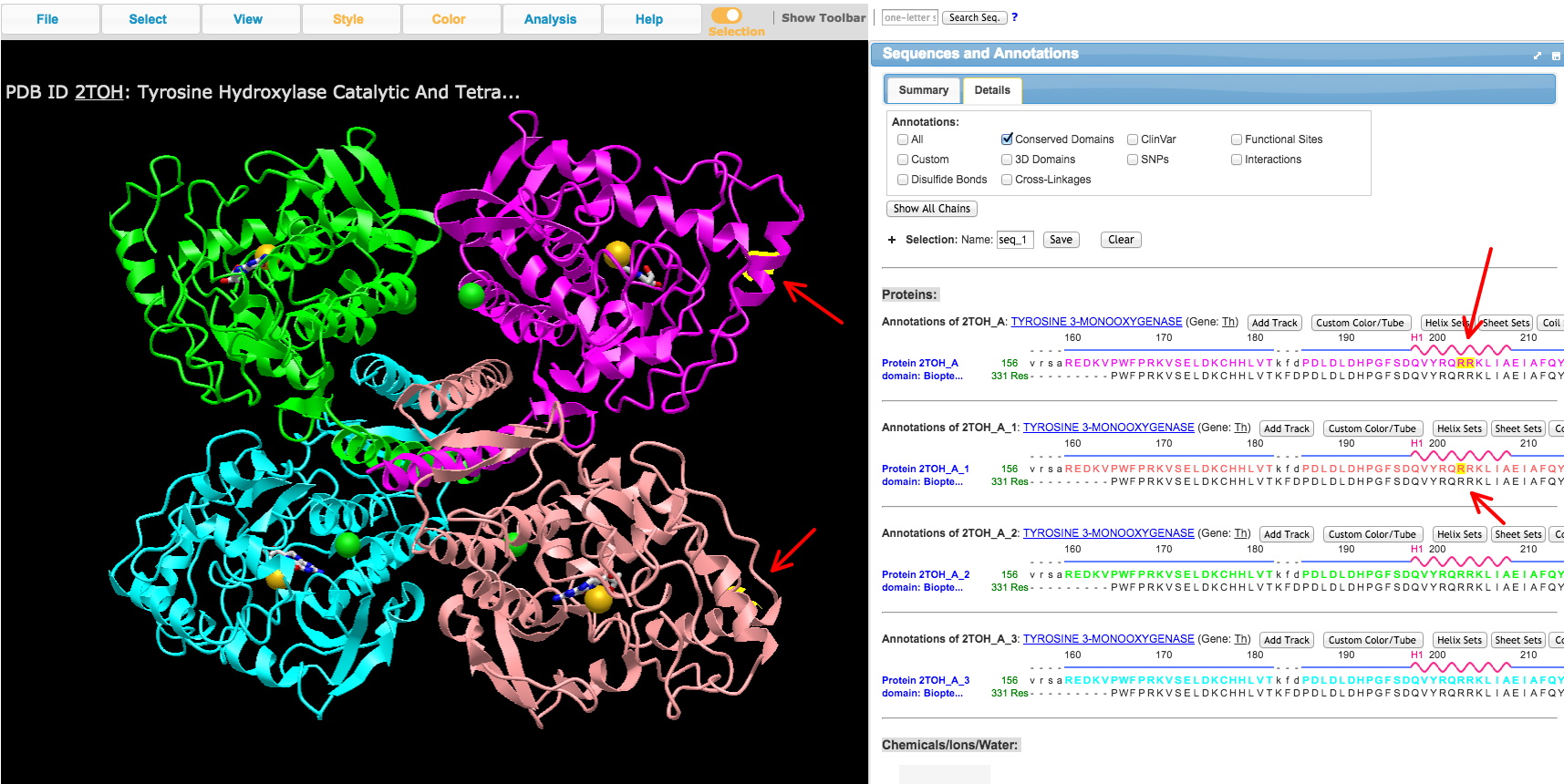
a. Under the Analysis menu, select 'View Sequences & Annotations,' and then select the 'Details' tab to see the AA sequence alongside the structure.
b. Select an amino acid or range of amino acids in the primary sequence to highlight the location in the structure. You can identify domains of the protein you may have read about from literature on your TF of interest.
c. Take a screenshot of the 3D protein structure you viewed in iCn3D for each domain of interest you've identified, especially the DNA-binding domain, with the AAs of the domain selected to turn in. Include the part of the right side window with the AA sequence showing the AAs selected. An example is shown below. In this case, the AA (R) was selected in two copies in the sequence window to show up on 2 of the 3D protein chains of the tetrameric crystal structure. The program highlights the location in yellow.